Pseudosection explorers tutorial
psexplorers provides several post-processing methods and visualizations of already constructed pseudosections. You can also create isopleths diagrams or do calculations along paths.
It provides four command-line scipts psgrid, psshow and psiso for quick visualizations. For options, check built-in help, e.g.:
$ psshow -h
To show pseudosection, you need to provide project file:
$ psshow -b /some/path/project.ptb
Using psexplorers in Python (or Jupyter notebook)
To access all options of psexplorer it is suggested to use Python API. You need to activate the pyps environment and run python interpreter:
$ conda activate pyps
$ python
To use psexplorer we need to import appropriate class, which contains most of the methods to work with pseudosection.
PTPSclass for P-T pseudosection constructed withptbuilder
TXPSclass for T-X pseudosection constructed withtxbuilder
PXPSclass for P-X pseudosection constructed withpxbuilder
All following commands must be executed in Python interpreter.
from pypsbuilder import PTPS
The second step is to create instance of pseudosection using existing project file.
pt = PTPS('/some/path/project.ptb')
We can check, whether the pseudosection already contains gridded
calculations. If not, we can use calculate_composition method to
calculate compositional variations on grid. The resulting data are stored in
project file. Note that any new modifications of the project by pypsbuilder*
will discard compositional variations on grid and must be calculated again.
if not pt.gridded:
pt.calculate_composition(nx=50, ny=50)
Gridding 1/1: 100%|██████████| 2500/2500 [03:10<00:00, 13.12it/s]
Grid search done. 0 empty points left.
Visualize pseudosection
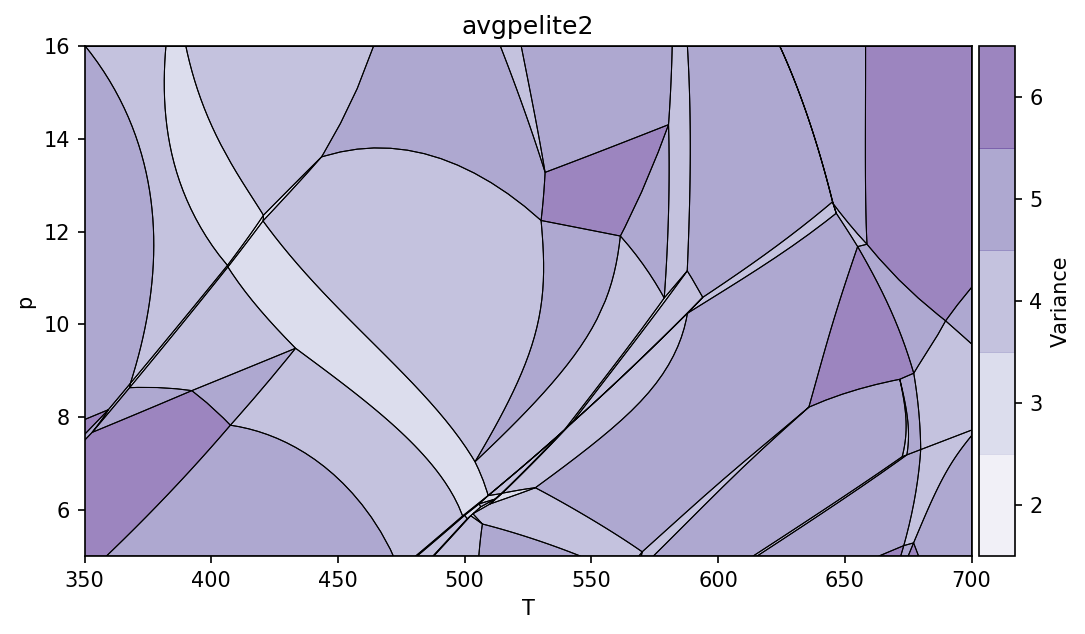
To show pseudosection, we can use show method
pt.show()
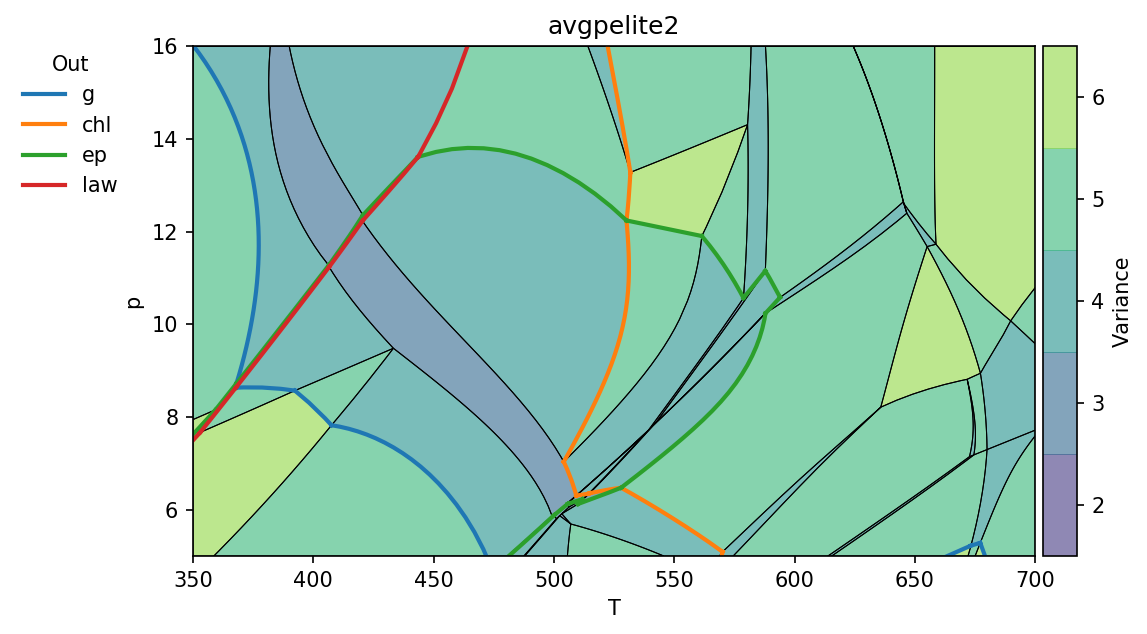
The keyword arguments cmap and out could be used to modify colormap and
highlight zero mode lines across pseudosection.
pt.show(cmap='viridis', out=['g', 'chl', 'ep', 'law'])
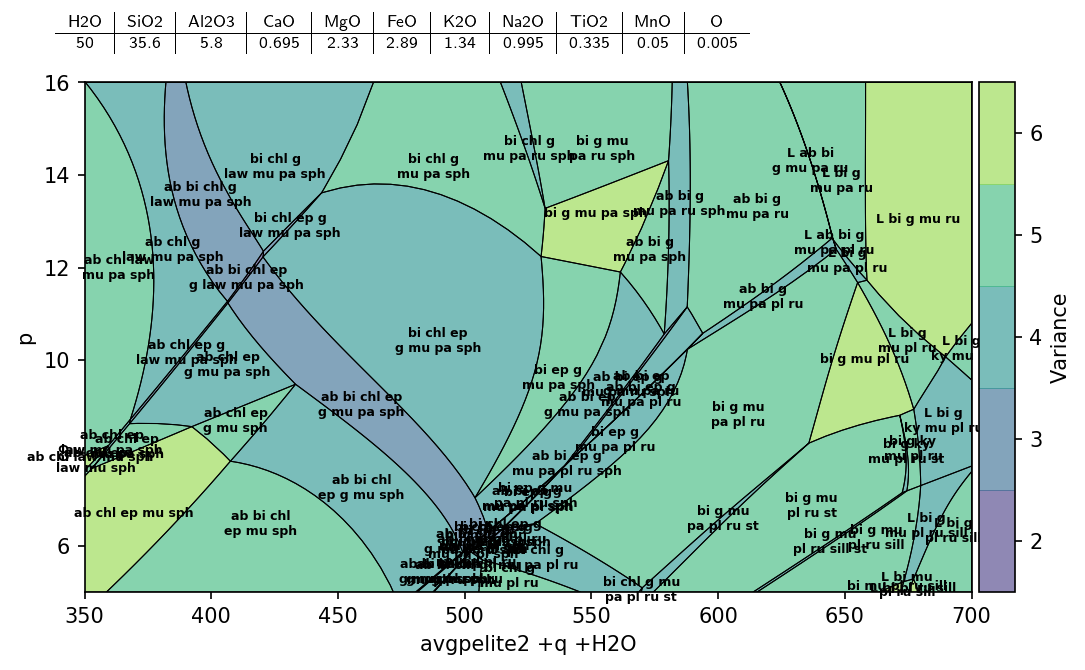
The keyword arguments bulk and label set whether the bulk composition
is shown on figure and whether the fields are labeled by phases.
pt.show(cmap='viridis', bulk=True, label=True)
The pseudosection identify method could be used to identify stable assemblage for
given p and T conditions. Note that returned key (Python frozenset) is used
to identify stable assemblage in many PTPS methods.
key = pt.identify(550, 13)
print(key)
frozenset({'sph', 'pa', 'q', 'g', 'mu', 'H2O', 'bi'})
Access data and variables stored in project
The calculated data are usually accessed using stable assemblage key (see above).
Theera are three groups of data stored 1) at invariant points, 2) along univariant
lines and 3) on grid covering multivariate fields. To see data coverage and all
available variables, you can use show_data method. When no variable (or expression)
is provided, method will show available variables.
pt.show_data(key, 'g')
Missing expression argument. Available variables for phase g are:
mode x z m f xMgX xFeX xMnX xCaX xAlY xFe3Y H2O SiO2 Al2O3 CaO MgO FeO K2O Na2O TiO2 MnO O factor G H S V rho
Available end-members for g: kho gr alm py spss
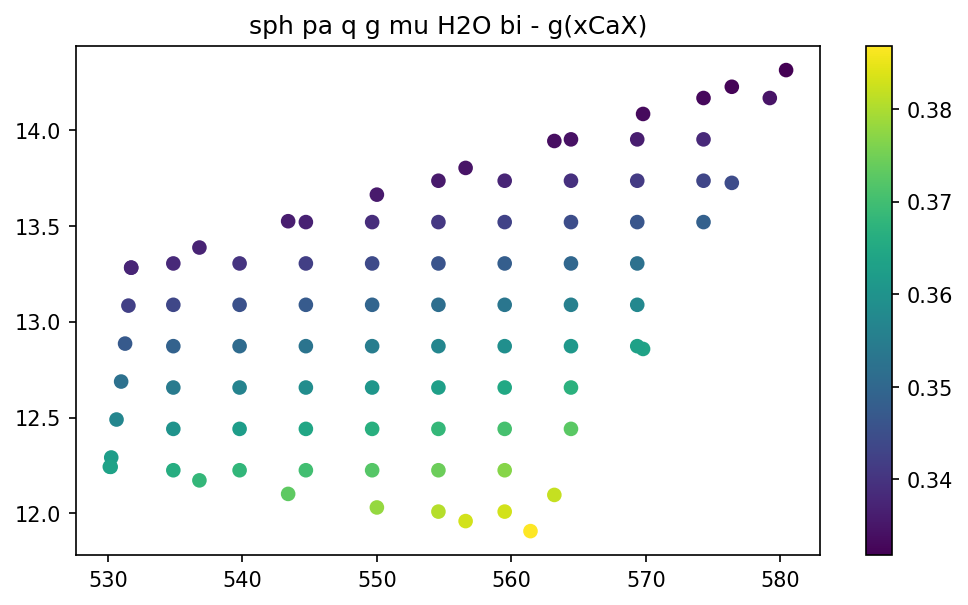
Once variable is provided, the all available data are shown.
pt.show_data(key, 'g', 'xCaX')
For data on the grid you can visualize them for all diagram in once using
show_grid method.
pt.show_grid('g', 'xCaX')
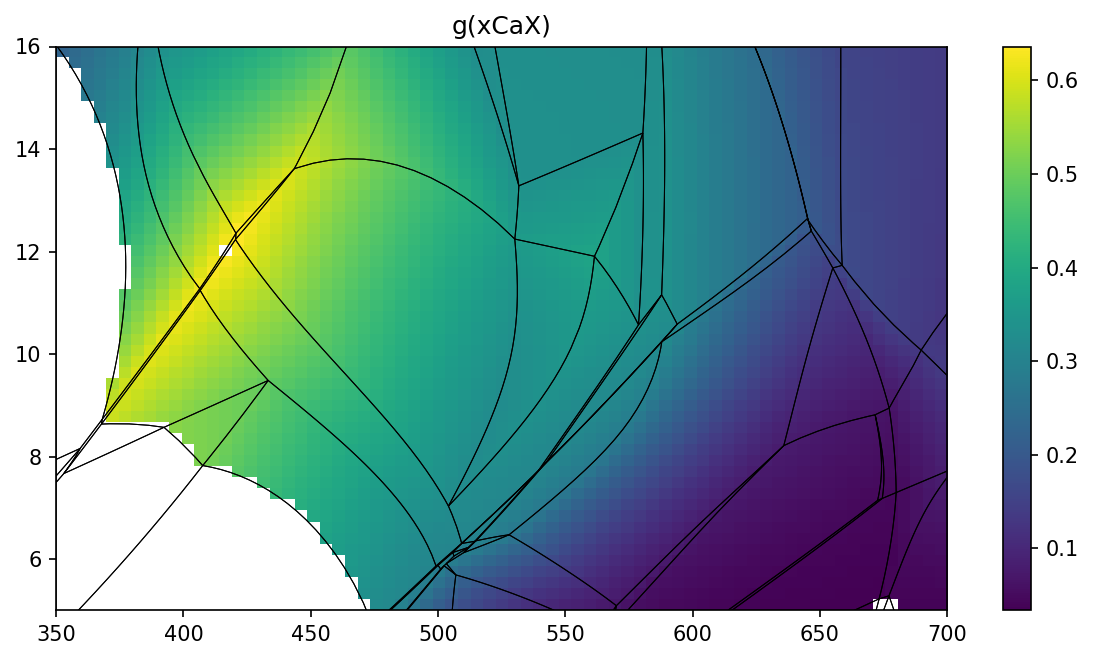
To create isopleths diagram you can use isopleths method. Note that
contours are created separately for each stable assemblage allowing
proper geometry of isopleths.
pt.isopleths('g', 'xCaX', N=14)
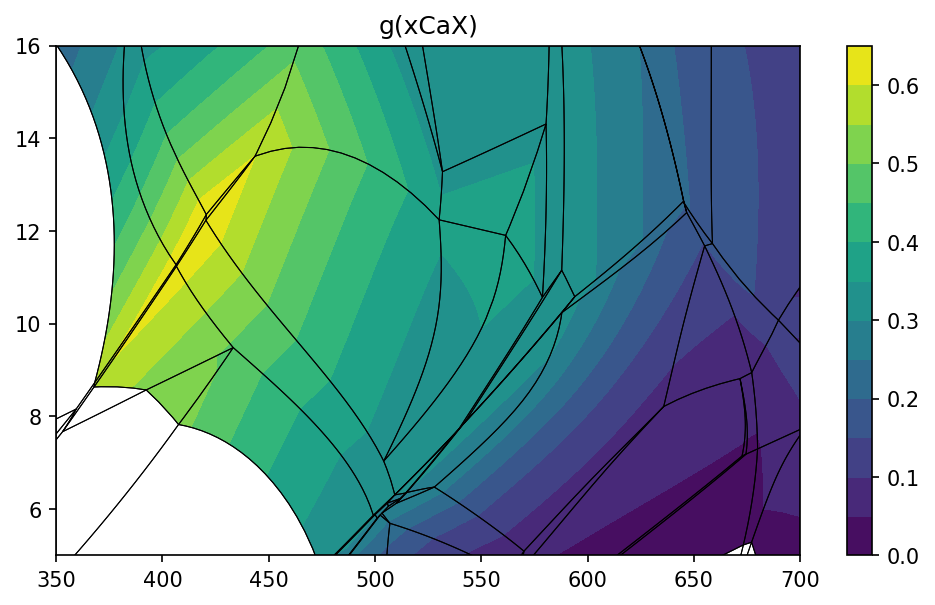
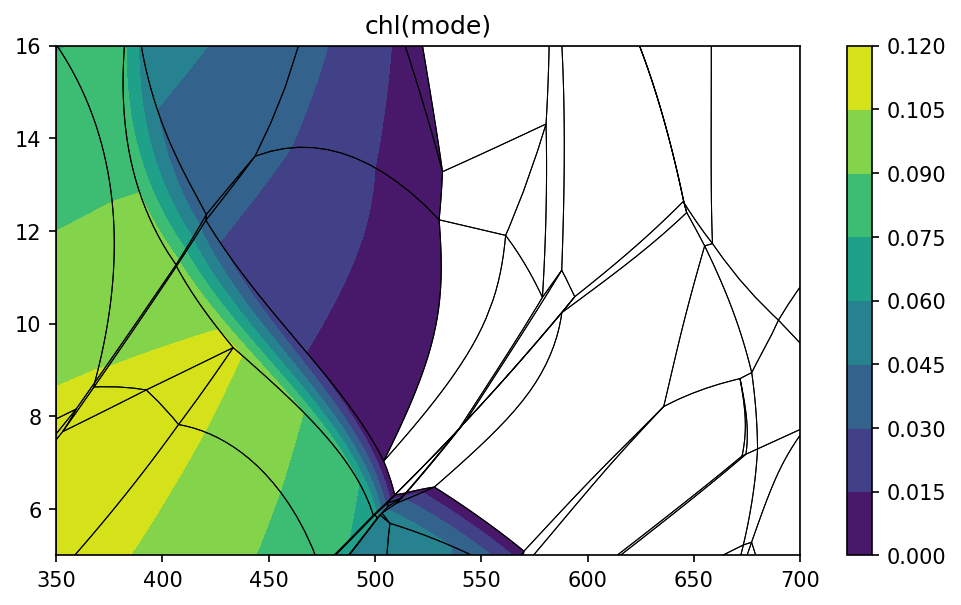
pt.isopleths('chl')
Missing expression argument. Available variables for phase chl are:
mode x y f m QAl Q1 Q4 xMgM1 xMnM1 xFeM1 xAlM1 xMgM23 xMnM23 xFeM23 xMgM4 xFeM4 xFe3M4 xAlM4 xSiT2 xAlT2 H2O SiO2 Al2O3 CaO MgO FeO K2O Na2O TiO2 MnO O factor G H S V rho
Available end-members for chl: ames mmchl ochl1 f3clin afchl ochl4 clin daph
pt.isopleths('chl', 'mode')
Calculations along PT paths
PTPS allows you to evaluate equilibria along user-defined PT
path. PT path is defined by series of points (path is interpolated) and
method collect_ptpath do actual calculations. It runs THERMOCALC
with ptguesses obtained from existing calculations.
t = [380, 480, 580, 640, 500]
p = [7, 12, 15, 9, 5.5]
pa = pt.collect_ptpath(t, p)
Calculating: 100%|██████████| 100/100 [00:03<00:00, 25.86it/s]
You can see phase modes along PT path using show_path_modes method.
pt.show_path_modes(pa, exclude=['H2O'])

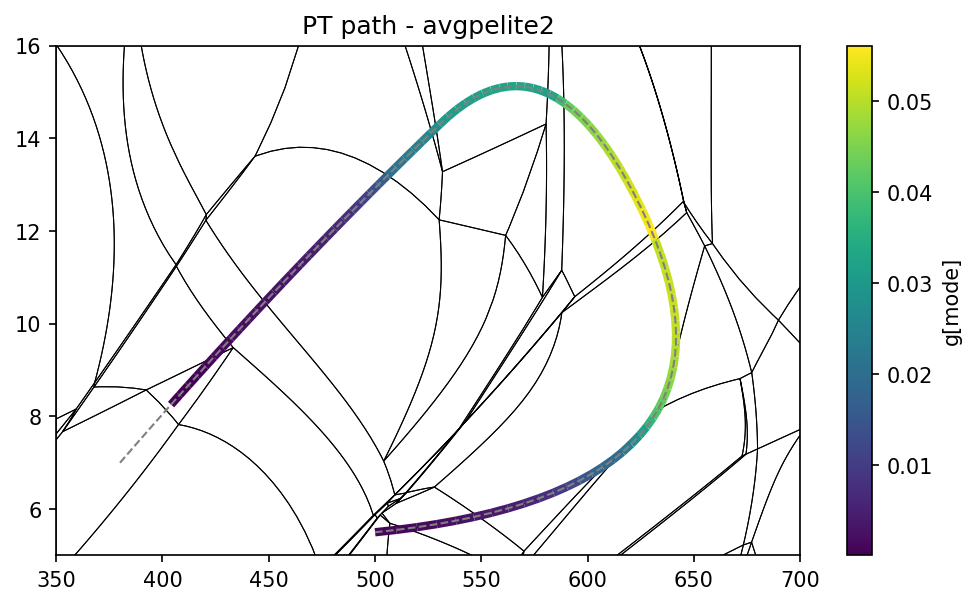
or show value of user-defined expression shown as colored strip on PT space.
pt.show_path_data(pa, 'g', 'mode')
Extra
show_status method shows status of calculations on the grid.
Possible failed calculations are shown.
pt.show_status()
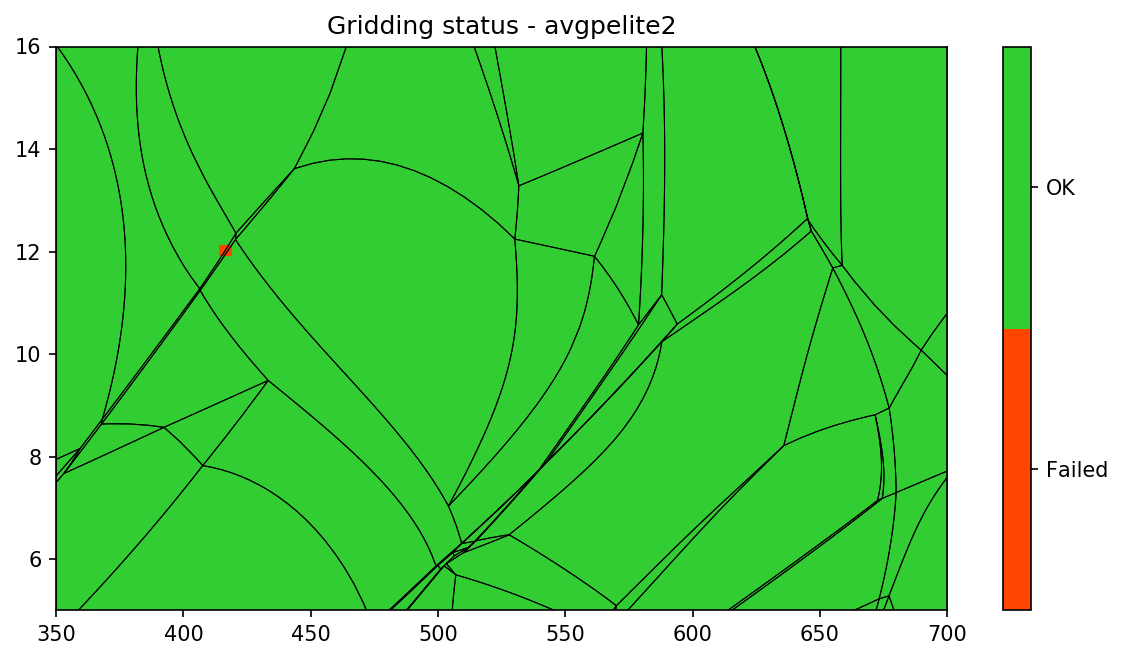
Do you want to know execution time of THERMOCALC on individual grid
points? Check show_delta method.
pt.show_delta(pointsec=True)
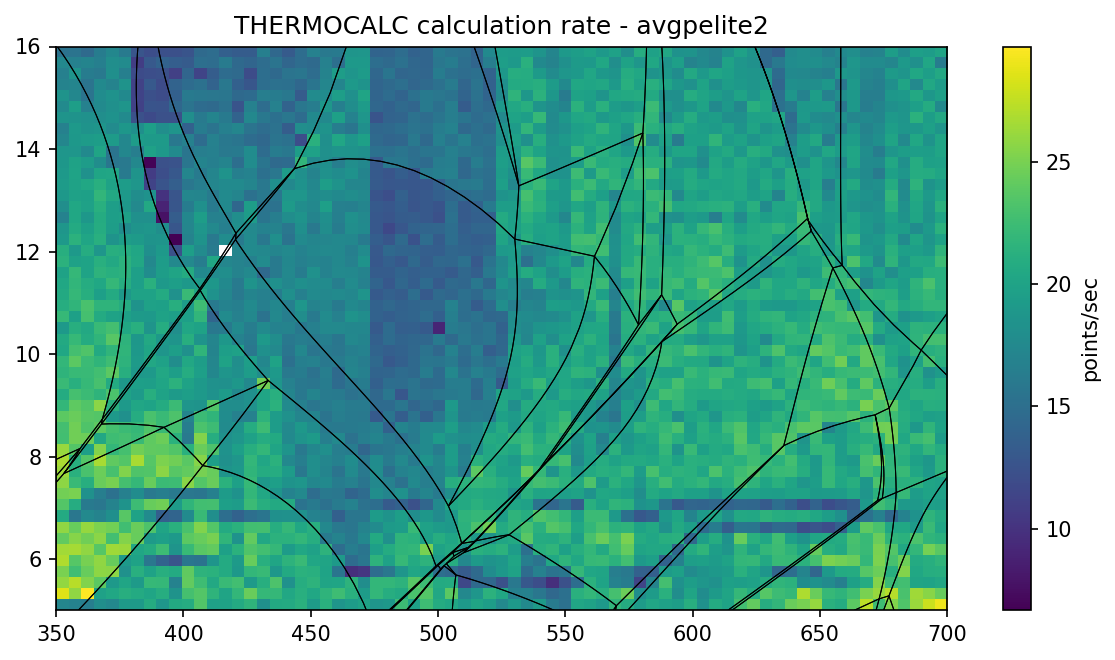
For full description of Python API check: Python API.